Research
In the Raj lab, we develop novel probes, biosensors, molecular imaging agents, and new chemoselective reactions for global profiling of undruggable proteome, selective detection of lysine posttranslational modifications (PTMs), and the synthesis of mechanically interlocked structures, cyclic peptides, and bioconjugates. This research program leads to the discovery of novel protein biomarkers, enzyme inhibitors, affordable diagnostic tools for early detection of cancer, and endogenous protein partners thus facilitating the synthesis of biotherapeutics. We utilize organic chemistry tools, biochemical techniques, and structural characterization.
Project
We are broadly interested in the dynamic networks of protein–protein and protein–nucleic acid interactions that underlie cellular function. In particular, our research focuses on identifying and characterizing covalent and non-covalent crosslinks between these biomolecules, with an emphasis on how these interactions are perturbed in disease states such as cancer, neurodegeneration, and viral infection.
By leveraging advanced chemical biology tools, crosslinking mass spectrometry (XL-MS), and systems-level proteomics, we aim to map interaction networks with high specificity and resolution. Understanding how interaction partners shift, disappear, or emerge under pathological conditions provides critical insights into disease mechanisms that cannot be gleaned from gene or protein expression alone.
Proteins that exhibit altered interaction profiles in disease represent promising nodes for therapeutic intervention—either as direct drug targets or as biomarkers for diagnosis and monitoring. Our work in interactomics thus not only deepens our understanding of cellular complexity but also opens the door to novel strategies for therapeutic discovery.
Selection of Recent Papers
- Wang, Y.; Czabala, P.; Raj, M. Bioinspired one-pot furan-thiol-amine multicomponent reaction for making heterocycles and its applications. Nat. Commun. 2023, 14, 4086-4101.
Project
We are interested in developing novel methods for the chemoselective modification of peptides, with the goal of expanding their chemical diversity and enhancing their therapeutic potential. Peptides offer a unique scaffold for drug development due to their biological specificity and modular architecture, but their utility is often limited by poor pharmacokinetic properties, low membrane permeability, and rapid degradation.
To overcome these challenges, we are designing late-stage functionalization (LSF) strategies that allow for the precise installation of new chemical motifs onto fully assembled peptide backbones. These methods enable rapid diversification of peptide libraries without the need to resynthesize the core sequence, dramatically accelerating structure–activity relationship (SAR) studies.
Our approaches focus on site-selective reactions that target underutilized amino acid residues (e.g., methionine, arginine, histidine), allowing for controlled modification without affecting the peptide’s biological function. These modifications can endow peptides with a range of customized physical and biological properties, including enhanced cytotoxicity, selective organelle targeting, improved proteolytic stability, and incorporation of imaging moieties such as fluorophores or photoaffinity tags.
By merging principles of medicinal chemistry with advanced bioconjugation techniques, our lab aims to develop functionally tailored peptides capable of reaching previously inaccessible therapeutic targets, including intracellular proteins and protein–protein interaction interfaces. This work ultimately supports the discovery of more effective, selective, and tunable peptide-based therapeutics.
Selection of Recent Papers
- Villalobos Galindo, A.; Raj, M. Solvent-Dependent Chemoselectivity Switch to Arg-Lys Imidazole Cross-Links. Org. Lett. 2024, 26, 8356.
- Emenike, B.*; Shahin, S.*; Raj, M. Bioinspired Synthesis of Allysine for Late-Stage Functionalization of Peptides. Angew. Chem., Int. Ed. 2024 e202403215.
- Prosser, L. P.*; Talbott, J. M.*; Garrity, R. P.; Raj, M. C-Terminal Arginine-Selective Cleavage of Peptides as a method for Mimicking Carboxypeptidase B. Org. Lett. 2023, 25, 6206-6210.
Project
The ability to selectively modify biomolecules is essential for probing biological systems and discovering disease biomarkers. Our lab develops novel bioconjugation methods that enable chemoselective labeling of proteins under physiological conditions. We focus on residues that are historically underutilized in chemical biology—such as methionine, arginine, and histidine—expanding the chemical space for selective targeting.
We have previously developed site-selective methods for lysine mono- and di-methylation as well as histidine methylation. These tools allow us to install functional groups that support detection, enrichment, or modulation of protein function, especially in the context of post-translational modifications (PTMs). Given the central role of PTMs in regulating cellular processes—and their dysregulation in disease—our work provides powerful new ways to study biological signaling and pathology.
We apply these chemical tools in both cell-based systems, animal models, and human clinical samples to uncover new disease-associated modifications, discover biomarkers, and better understand the underlying molecular mechanisms of human health and disease.
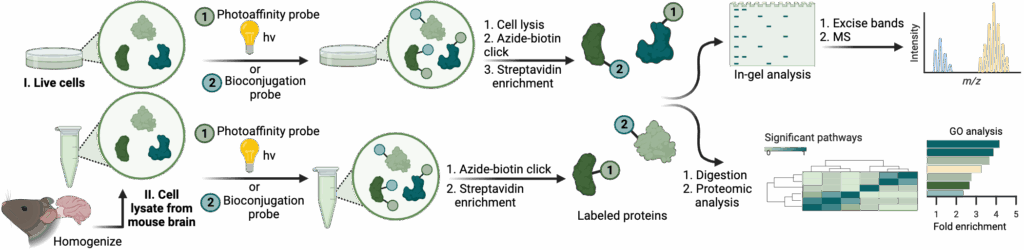
Selection of Recent Papers
- Paikin, Z. E.*; Emenike, B.*; Shirke, R.; Beusch, C. M.; Gordon, D. E.; Raj, M. Acrolein-Mediated Conversion of Lysine to Electrophilic Heterocycles for Protein Diversification and Toxicity Profiling. J. Am. Chem. Soc. 2025, 147, 5679.
- Sahu, S.*; Emenike, B.*; Buesch, C. M.; Bagchi, P.; Gordon, D. E.; Raj, M. Copper(I)-nitrene platform for chemoproteomic profiling of methionine. Nat. Commun. 2024, 15, 4243.
- Emenike, B.; Czabala, P.; Farhi, J.; Swaminathan, J.; Anslyn, E. V.; Spangle, J.; Raj, M. Tertiary Amine Coupling by Oxidation for Selective Labelling of Dimethyl Lysine Post-Translational Modifications. J. Am. Chem. Soc. 2024, 146, 10621-10631.
Project
The intersection of chemistry and data science holds enormous potential to transform therapeutic discovery. Our lab is developing machine learning (ML) and artificial intelligence (AI) models to enhance prediction, screening, and optimization in chemical biology.
One of our current efforts focuses on using machine learning to predict the cell permeability of peptides—a major limitation in peptide drug development. By integrating experimental data from permeability assays with peptide sequence and structural information, we aim to build predictive models that can guide the design of more effective therapeutic peptides.
We are also interested in applying AI to other areas of our research, such as analyzing complex proteomic datasets, modeling protein interaction networks, and optimizing sensor design. These data-driven approaches allow us to accelerate hypothesis generation, identify patterns that may not be obvious through traditional analysis, and design better experiments.
Selection of Recent Papers
- Bruce, A.*; Adebomi, V.*; Czabala, P.; Palmer, J.; McFadden, W. M.; Lorson, Z. C.; Slack, R. L.; Bhardway, G.; Sarafianos, S. G.; Raj, M. A Tag-Free Platform for Synthesis and Screening of Cyclic Peptide Libraries. Angew. Chem., Int. Ed. 2024, 63, e202320045.
- Shao, H.; Adebomi, V.; Bruce, A.; Raj, M.; Houk, K. N. Intramolecular Hydrogen Bonding Enables a Zwitterionic Mechanism for Macrocyclic Peptide Formation: Computational Mechanistic Studies of CyClick Chemistry. Angew. Chem., Int. Ed. 2023, e202307210.
Project
Aldehydes are classified as cytotoxic because they damage DNA and proteins by forming interstrand cross-link in human cells. The human body tightly regulates the concentration of aldehydes by aldehyde dehydrogenases that convert toxic aldehydes to nontoxic acids. The mutation in the enzyme ALDH2*2 (present in approximately ~ 0.6 billion people worldwide) leads to accumulation of toxic aldehydes thus contribute to a variety of human diseases including cardiovascular diseases, diabetes, neurodegenerative diseases, stroke, and cancer. Despite the cytotoxicity associated with aldehydes, there are no pharmaceuticals available to eradicate these molecules. This is largely due to the lack of methods to measure the levels of multiple alkyl aldehydes in cells. The main goal of this research proposal is to fill the present gap in the range of available techniques to measure aldehyde levels in cells and the development of new therapeutics. We seek to develop a new family of chemical sensors to measure the total aldehyde levels inside the cell and the concentrations of individual aldehydes. Furthermore, we have begun applying our sensors into mice models to visualize aldehydes in relation to neurodegenerative diseases.
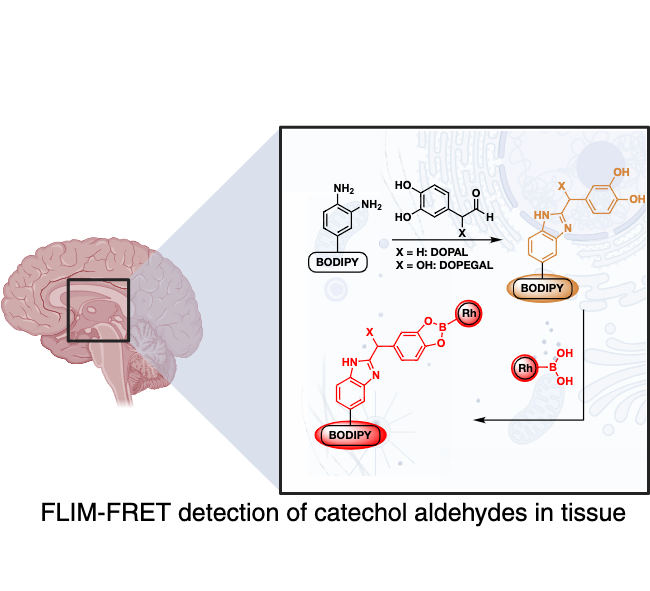
Selection of Recent Papers
- Wills, R.*; Shirke, R.*; Hrncir, H.; Talbott, J. M.; Sad, K.; Spangle, J. M.; Gracz, A. D.; Raj, M. Tunable fluorescent probes for detecting aldehydes in living systems. Chem. Sci. 2024, 15, 4673-4769.
- Wills, R.; Farhi, J.; Czabala, P.; Shahin, S.; Spangle, J. M.; Raj, M. Chemical sensors for imaging total cellular aliphatic aldehydes in live cells. Chem. Sci. 2023, 14, 8305-8314.
Project
We are committed to exploring next-generation therapeutic strategies that go beyond traditional small molecules. This includes both new molecular modalities and novel approaches to targeting disease mechanisms.
One major area of interest is the development of peptide-based inhibitors and cyclic peptide libraries to target proteins that are traditionally considered “undruggable.” Using tools like CyClick, we are able to generate diverse, stable, and bioactive macrocycles that can modulate challenging protein–protein interactions.
We are also working on platforms to explore PROTACs and molecular glues for targeting currently untreatable disease states.
Our long-term goal is to develop conditionally active therapeutics, tissue-targeted delivery systems, and multifunctional agents that integrate sensing, imaging, and therapeutic action. This integrative approach merges chemistry, biology, and engineering to pioneer innovative solutions to unmet medical needs.
Selection of Recent Papers
- Bruce, A.*; Adebomi, V.*; Czabala, P.; Palmer, J.; McFadden, W. M.; Lorson, Z. C.; Slack, R. L.; Bhardway, G.; Sarafianos, S. G.; Raj, M. A Tag-Free Platform for Synthesis and Screening of Cyclic Peptide Libraries. Angew. Chem., Int. Ed. 2024, 63, e202320045.
- Shao, H.; Adebomi, V.; Bruce, A.; Raj, M.; Houk, K. N. Intramolecular Hydrogen Bonding Enables a Zwitterionic Mechanism for Macrocyclic Peptide Formation: Computational Mechanistic Studies of CyClick Chemistry. Angew. Chem., Int. Ed. 2023, e202307210.